The task for this activity is to enhanced an image using histogram manipulation.
For this activity, we are asked to find grayscale images. I looked for an image in the web. The image is from: http://upload.wikimedia.org/wikipedia/commons/thumb/9/90/Face_sapa14.jpg/800px-Face_sapa14.jpg
To find the histogram of the image:
Change the directory where the image is saved.
img = imread('face.jpg');
// plot histogram
gsval=[];
pixelnum=[];
counter=1;
for i=0:1:255
[x,y]=find(img==i);
gsval(counter)=i;
pixelnum(counter)=length(x);
counter = counter+1;
end;
scf(1);
plot(gsval,pixelnum);
// puts the titles
xtitle( 'Histogram') ;
This will show
Note: x-axis is the grayscale values and the y-axis is the number of pixels.
It ii show in the histogram that the grayscale values is well spread in the image.
After finding the histogram, we can find the probability distribution function (PDF). PDF can be find by normalizing the pixel values of the histogram. After finding the PDF, we will find the cumulative distribution function (CDF).
To find the PDF (normalized histogram) and CDF
//plot PDF
a = gsval;
b = pixelnum;
c = size(img,1);
d = size(img,2);
e = c.*d
f = b/e;
scf(2);
plot(a,f);
g = cumsum(f);
xtitle ('Normalized Histogram') Note: x-axis is the grayscale image and the y-axis is the normalized number of pixels.
Note: x-axis is the grayscale image and the y-axis is the normalized number of pixels.
//plot CDF
scf(3);
plot(a,g);
xtitle('CDF');
Note: x-axis is the grayscale values while y-axis is the cumulative sum of the normalized number of pixels.
________________________________________________________
Then the next thing to do is the Back Projection of the Image
//Back Projection
gray = [];
for ii = 1:1:size(img,1)
for jj = 1:1:size(img,2)
gray(ii,jj) = g(img(ii,jj));
end
end
imwrite(gray, 'gray.jpg');
After back projection,

Then we will find the histogram and the CDF of the new image.
// New Histogram
im = imread('gray.jpg');
gval=[];
pnum=[];
count=1;
for i=0:1:255
[x1,y1]=find(im==i);
gval(count)=i;
pnum(count)=length(x1);
count = count+1;
end;
scf(4);
plot(gval,pnum);
xtitle('Histogram'); Note: x-axis is the grayscale values and the y-axis is the number of pixels.
Note: x-axis is the grayscale values and the y-axis is the number of pixels.
// New PDF and CDF
//plot PDF
h = gval;
i = pnum;
j = size(im,1);
k = size(im,2);
l = j.*k;
m = i/l;
plot(h,m);
xtitle('Normalized Histogram')
Note: x-axis is the grayscale image and the y-axis is the normalized number of pixels.
//plot new CDF
n= cumsum(m);
scf(5);
plot(h,n);
xtitle(' CDF');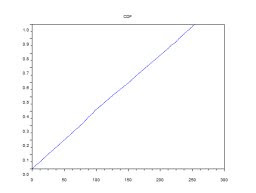 Note: x-axis is the grayscale values while y-axis is the cumulative sum of the normalized number of pixels.
Note: x-axis is the grayscale values while y-axis is the cumulative sum of the normalized number of pixels.
After finding the PDF and CDF of the new image. These plots show that the new image has a good contrast.
_________________________________________________________
The next task is to remap the CDF of the grayscale image to find the desired CDF.

 I used the function
I used the function
G = 0.5*[tanh(6*((Z-128)/255))+1] where Z = [0:255]
The graph of this function is our desired CDF Applying the function by interpolation.
Applying the function by interpolation.
Z = [0:255];
G = tanh(6.*(Z-128)./255);
G = (G+1)/2;
A = interp1(a,g,img);
graynew = interp1(G,Z,A);
imshow(graynew, [0 255]);
The final image is We then find the histogram, PDF and CDF using same commands we used in finding the histogram, PDF and CDF of the original image.
We then find the histogram, PDF and CDF using same commands we used in finding the histogram, PDF and CDF of the original image.
Histogram: Note: x-axis is the grayscale values and the y-axis is the number of pixels.
Note: x-axis is the grayscale values and the y-axis is the number of pixels.
PDF: Note: x-axis is the grayscale image and the y-axis is the normalized number of pixels.
Note: x-axis is the grayscale image and the y-axis is the normalized number of pixels.
CDF: Note: x-axis is the grayscale values while y-axis is the cumulative sum of the normalized number of pixels.
Note: x-axis is the grayscale values while y-axis is the cumulative sum of the normalized number of pixels.
Comparing the three images:


The last image is darker than the other two but I think that the middle image has the best contrast. This is also based on the PDF and CDF of the images.
Acknowledgements:
Julie D.
Ed
Beth
Angel
Aiyin
for helping me and answering my questions.
Grade :
10/10, because I did my best and I enhanced the original image.
Thursday, June 26, 2008
A4 – Enhancement by Histogram Manipulation
Posted by
anarica
at
8:55 AM
0
comments
![]()
Tuesday, June 24, 2008
A3 - Image Types and Basic Image Enhancement
PART 1
There are 4 types of images
1. True Color Image
photo from: www.cssnz.org/flower.jpg
Using the command in Scilab
--> imfinfo ('flower1.jpg')
shows the following results
| File Name | flower1.jpg |
| File Size | 3348 |
| File Format | jpeg |
| Width | 115 |
| Height | 118 |
| Depth | 8 |
| Storage Type | truecolor |
| Number of Colors | 0 |
| Resolution Unit | centimeter |
| X Resolution | 0 |
| Y Resolution | 0 |
Using the properties in Windows
| Width | 115 |
| Height | 118 |
| Horizontal Resolution | 96 dpi |
| Vertical Resolution | 96 dpi |
| Bit Depth | 24 |
2. Gray Scale image
 photo from: digital-photography-school.com
photo from: digital-photography-school.comUsing the command in Scilab
--> imfinfo ('flower2.jpg')
shows the following results
| File Name | flower2.jpg |
| File Size | 4225 |
| File Format | JPEG |
| Width | 120 |
| Height | 120 |
| Depth | 8 |
| Storage Type | indexed |
| Number of Colors | 256 |
| Resolution Unit | inch |
| X Resolution | 76 |
| Y Resolution | 76 |
Using the properties in Windows
| Width | 120 |
| Height | 120 |
| Horizontal Resolution | 72 dpi |
| Vertical Resolution | 72 dpi |
| Bit Depth | 8 |
3.Indexed Image
 photo from:www.digitalphotoartistry.com/rose1.jpg
photo from:www.digitalphotoartistry.com/rose1.jpgUsing the command in Scilab
--> imfinfo ('flower3.bmp')
shows the following results
| File Name | flower3.bmp |
| File Size | 16306 |
| File Format | bmp |
| Width | 106 |
| Height | 141 |
| Depth | 8 |
| Storage Type | indexed |
| Number of Colors | 256 |
| Resolution Unit | centimeter |
| X Resolution | 28.350000 |
| Y Resolution | 28.350000 |
Using the properties in Windows
| Width | 106 |
| Height | 141 |
| Bit Depth | 8 |
4. Binary Image
 photo from:www.botany.com
photo from:www.botany.comUsing the command in Scilab
--> imfinfo ('flower4.bmp')
shows the following results
| File Name | flower4.bmp |
| File Size | 1550 |
| File Format | bmp |
| Width | 124 |
| Height | 93 |
| Depth | 8 |
| Storage Type | indexed |
| Number of Colors | 2 |
| Resolution Unit | centimeter |
| X Resolution | 28.350000 |
| Y Resolution | 28.350000 |
Using the properties in Windows
| Width | 124 |
| Height | 93 |
| Bit Depth | 1 |
PART 2:
This part of the activity is concerned with our scanned image.
Cambodian money( c/o Anthony Amarra)
 I cropped the image so that the rulers will not be included in reading the area of the scanned image.
I cropped the image so that the rulers will not be included in reading the area of the scanned image.
Using
-->image = imread('gray2.jpg');
-->imfinfo('gray2.jpg')
shows the following properties
FileName: gray2.jpg
Format: JPEG
Width: 422
Height: 201
Depth: 8
StorageType: indexed
NumberOfColors: 256
ResolutionUnit: inch
XResolution: 96.000000
YResolution: 96.000000
Using the command
-->histplot ([0:1:255], image)
We find the histogram of the image
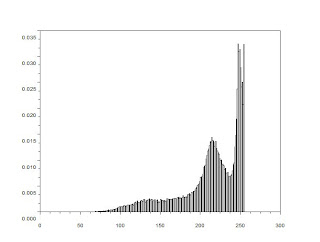 From the histogram we can say that the image is low in contrast.
From the histogram we can say that the image is low in contrast.We can also check the histogram and threshold by using imageJ.
 We can also find the histogram using the following commands:
We can also find the histogram using the following commands:-->gsval=[]
-->pixelnum=[]
-->counter=1;
-->for i=0:1:255
-->[x,y]=find(image==i);
-->gsval(counter)=i;
-->pixelnum(counter)=length(x);
-->counter = counter+1; -->end;
-->plot(gsval,pixelnum);
This will show the image below:
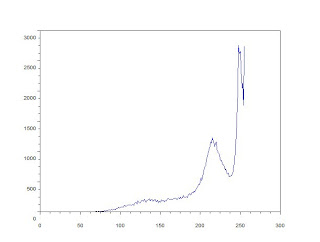
Using the histogram, i find the best threshold is at 189 or 74.117647% of 255. I used this threshold to convert the image to binary
-->bw = im2bw(image,0.74117647);
--> imshow(bw,2)
It shows the image below
 Inverting the image using the commands,
Inverting the image using the commands,--> binary = abs(bw-1); --> imshow (binary)
shows the image below:
 Using the commands, in the previous activity, to find the area
Using the commands, in the previous activity, to find the area-->[x,y] = follow(binary);
-->size(x)
ans =
924. 1.
-->size(y)
ans =
924. 1.
-->x_2(1) = x(924) ;
-->x_2(2:924) = x(1:923);
-->y_2(1) = y(924) ;
-->y_2(2:924) = y(1:923);
-->area=0.5*sum(x.*y_2-y.*x_2);
--> Area = abs(area)
Area =
84200.
The experimental value for area is 84200 square pixels.
The theoretical value for area is ( 419 x 199) 83381 square pixels. This can be solve by pixel counting
The percent error is 0.9822%.
Acknowledgments:
JULIE D.
ED
BETH
AIYIN
- for answering some of my questions
JORGE
- for the histplot
JERIC
- for the histogram code
GRADE:
10/10 because I did my best in this activity and the error is acceptable.
Posted by
anarica
at
11:05 AM
0
comments
![]()
Thursday, June 19, 2008
A2 = Area estimation of images
Using the sip toolbox in the scilab program, we have can view images.
We open the scilab program then we open the sip toolbox but we encounter error in the imagemagick path so we type command so that we can use sip toolbox.
The commands used are:
chdir(ImageMAgickPath); link(CORE_RL_Magick_.dll);
after entering this command, we can use the sip toolbox.
I first studied how to insert images in the sip toolbox. I first changed the directory using the command
chdir ('C:\Documents and Settings\AP186user13\My Documents\My Pictures').
After changing the directory, I can now read the image using sip tool box.
To read the image, I used the commands
-->image = imread('square.bmp');
-->xbasc(); imshow(image);
Then, the image is shown in the sip graphic window. This image(which is binary) will appear Now, i am ready to use "follow" and find the area of the image using scilab.
Now, i am ready to use "follow" and find the area of the image using scilab.
I enter the following commands to find the area of the square.
--> [x,y] = follow(image);
--> size(x);(the size of x is 584)
--> size(y); (the size of y is 584)
--> x2(1) = x(584) ;
--> x2(2:584) = x(1:583);
-->y2(1) = y(584) ;
-->y2(2:584) = y(1:583);
--> area=0.5*sum(x.*y2-y.*x2);
area = 21316.
The dimensions of the square is 148x148. Therefore the theoretical value of the square is 21904.
The error from the value at the scilab to the value of the theoretical is 2.69%.
For the second, rectangle Using the same commands as the square,
Using the same commands as the square,
-->image = imread('rectangle.bmp');
-->xbasc(); imshow(image);
--> [x,y] = follow(image);
--> size(x);(the size of x is 510)
--> size(y); (the size of y is 510)
--> x2(1) = x(510) ;
--> x2(2:510) = x(1:509);
-->y2(1) = y(510) ;
-->y2(2:510) = y(1:509);
--> area=0.5*sum(x.*y2-y.*x2);
area = 14534.
we find the scilab value of area to be 14534 and the theoretical to be 14960. Therefore the error is 2.84%.
For the third image, Using the same commands.
Using the same commands.
-->image = imread('circle.bmp');
-->xbasc(); imshow(image);
--> [x,y] = follow(image);
--> size(x);(the size of x is 548)
--> size(y); (the size of y is 548)
--> x2(1) = x(548) ;
--> x2(2:548) = x(1:547);
-->y2(1) = y(548) ;
-->y2(2:548) = y(1:547);
--> area=0.5*sum(x.*y2-y.*x2);
area = 29477
The scilab value of area is 29477. While the theoretical(having a radius of 97) is 29599. The error is 0.02%.
Acknowledgements...
Beth, for answering all my questions...
Lei, for helping me with the commands...
I will give myself a grade of 10/10.
Good day.
Posted by
anarica
at
10:58 AM
2
comments
![]()
Thursday, June 12, 2008
A1-Digital Scanning
Using the scanned handwritten graph found in the College of Science Library.
from
Title: Electrolytic Determinations & Separations with the Use of a Rotating Anode.
Reference: J. Langness, The Journal of the American Chemical Soc. 29, 459-472 (1907). 
 The second and third columns are the pixel locations of the data points. The fourth and fifth columns are the points when the pixel location of a data is subtracted to the pixel location of the origin. However, in the x-axis I use the absolute value.
The second and third columns are the pixel locations of the data points. The fourth and fifth columns are the points when the pixel location of a data is subtracted to the pixel location of the origin. However, in the x-axis I use the absolute value.The last two columns are the ratio of the x and y. We multiply the value of the major grid to the value of the pixel location and divide it by the pixel value. For example, data point 1(x-axis)
 Comparing the two graphs,
Comparing the two graphs,

I have found out that the graphs are almost the same. But because of human error of looking at the exact value of the data points, the graph will look like this.

I made some errors but I still did my best so I will give myself 10/10...
Credits to:
Elizabeth Ann Prieto for scanning the images and for answering some of my questions... :D
Good day.. :)
Posted by
anarica
at
9:04 AM
1 comments
![]()
